Join us
We are looking for strongly motivated students with an interest in applying machine learning to computer vision, robotics and more broadly artificial intelligence.
Internships and thesis projects can lead to a PhD in the ELLIS Unit at CIIRC in Prague with the possibility of spending part of the time at other Units in the ELLIS network.
Internship and Thesis topics
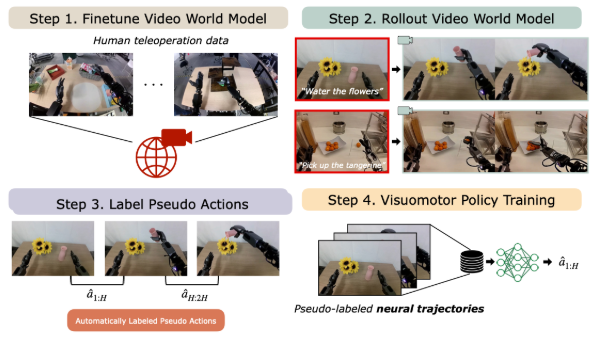
Inverse Dynamic Models for robotics
Supervisors
Georgij Ponimatkin, Vladimir Petrik, and Josef Sivic
Motivation
The goal of this project is to explore the design and performance of inverse dynamic models (IDM) available today. In its essence, IDM is a model that takes as input two robot observations (often frames generated by a world model [DreamGen]) and predicts an action between them. The action is in the form of a change in the robotic joint positions or robotic gripper position.
This is useful for either creating policies driven by world models or using world models as a way to generate novel training data on the fly for training of large robotic VLA models such as pi0 [Pi0]. Here, IDMs allow for extracting actions (such as a change in a robot joint angles) between two consecutive frames and use them either directly as an action for the robot to execute or use them as a label for training large robotic models.
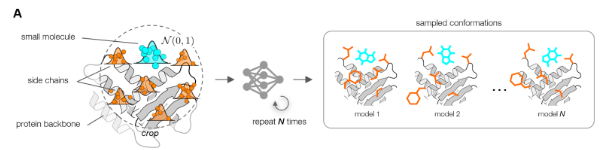
Customization of generative molecular docking models with quantum mechanical data
Supervisors
Jakub Kopko, Jiri Sedlar and Josef Sivic, in collaboration with researchers from Loschmidt Laboratories at Masaryk University in Brno (Saltuk Eyrilmez, David Bednar, and Stanislav Mazurenko)
Motivation
This project explores the use of modern AI models for molecular docking, such as PLACER (formerly ChemNet, Baker Lab) and 3DMolFormer. These models try to predict how a small molecule (a potential drug) might fit into a protein, like finding the right key for a lock — a crucial step in computer-aided drug design.
The goal is to customize these models for specific proteins using high-quality, in-house quantum mechanical data. By doing so, we aim to improve their ability to find promising molecules in virtual screening — a process where computers rapidly test millions of molecules to identify those most likely to become effective drugs, long before any laboratory experiments.
Learning Local Features from Generative Image Models
Supervisors
Torsten Sattler
Motivation
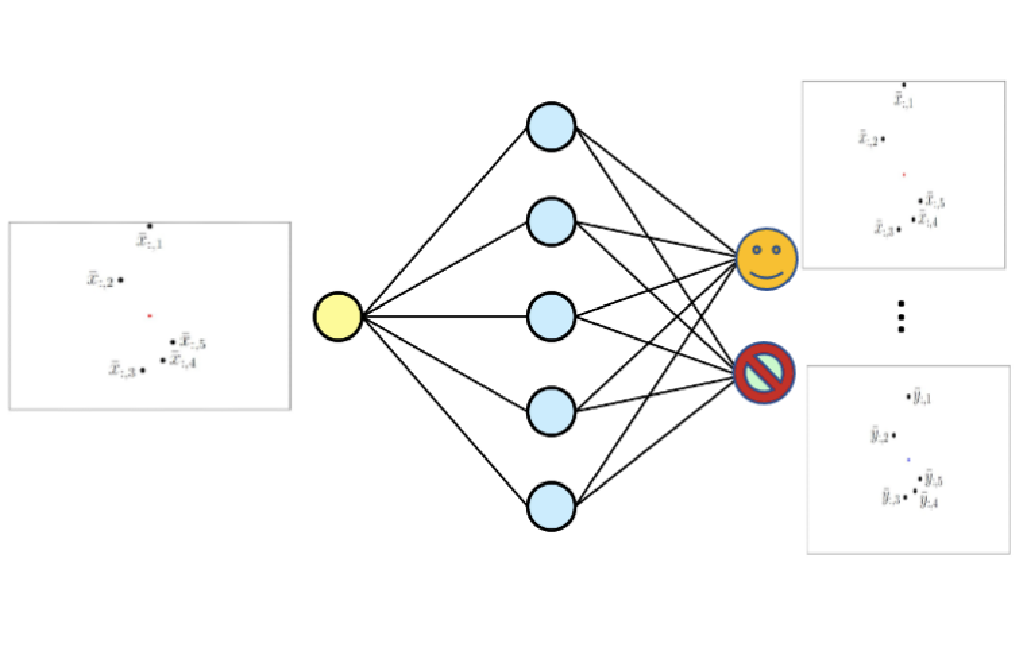
Local features play an important role in many 3D computer vision algorithms, including visual localization and Structure-from-Motion. A typical requirement of these algorithms is that features can be robustly matched under changing conditions, e.g., day-night changes or seasonal changes. However, acquiring suitable training data to train robust features is a challenging problem itself: First of all, it is necessary to obtain data of the same place under different conditions. Next, one needs to associate these images. The idea behind this thesis is to avoid these issues using modern generative models that can generate images based on text prompts, e.g., Stable Diffusion.
Learning to solve multiple-view geometry
in Computer Vision
Supervisors
Tomas Pajdla
Motivation
We aim at using machine learning to address long-standing problems in multiple view geometry that traditional techniques cannot solve. Previous methods for computing camera geometry from image matches can coped efficiently with only relatively simple problems in two-view geometry. We have developed first steps to solving hard problems in multiple-view geometry by using machine learning to tune techniques from numerical algebraic geometry to the data, thus making them tractable and efficient [LSHMP,FVGURD].
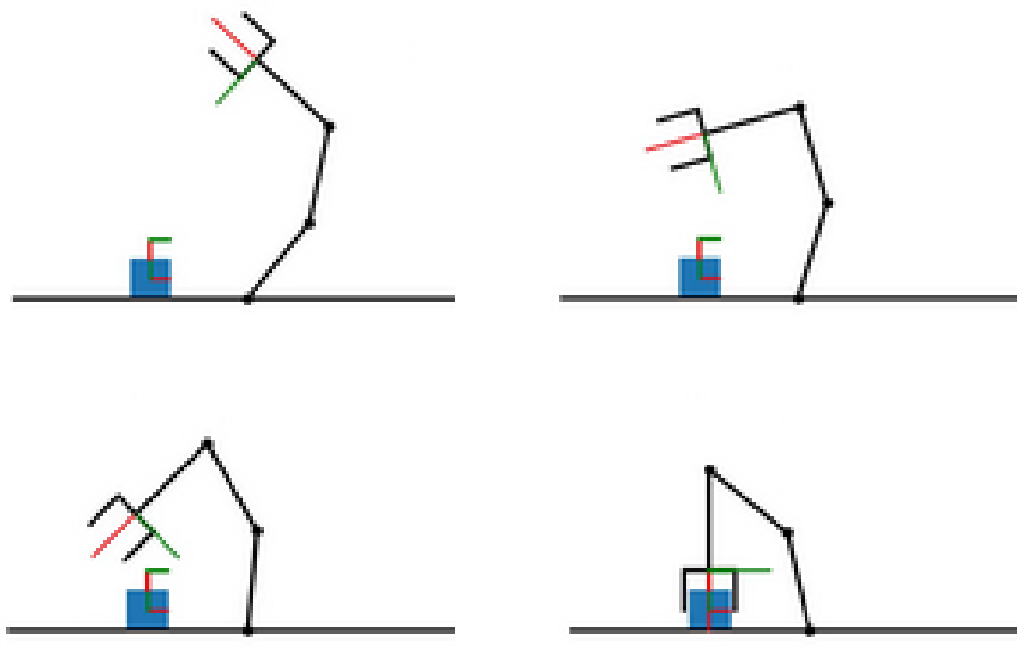
Generative models for robot motion representation
Supervisors
Mederic Fourmy, Vladimir Petrik, Josef Sivic
Motivation
The goal of this project is to use similar architecture to generate the motion of the robot given the prompt defined as the start and goal configuration of the robot and by encoded geometrical representation of the surrounding environment. The training dataset will be created by solving task-and-motion planning problems via Humanoid Path Planner. The goal is to study the generalization capability of the model to predict motions that were not seen during the training. The direct application of the motion representation is to warm-start the optimal control problem solver for robot control to avoid solving computationally expensive path planning problems online.
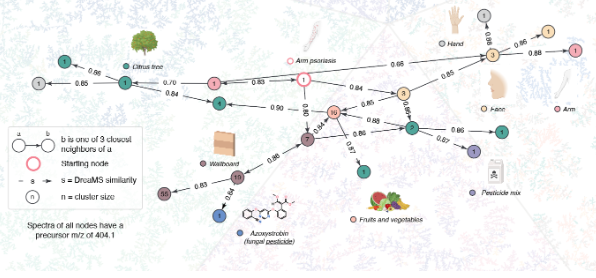
Web interface for exploring the chemical universe of DreaMS Atlas
Supervisors
Roman Bushuiev, Anton Bushuiev, Josef Sivic, Tomáš Pluskal
Motivation
Tandem mass spectrometry (MS/MS) is the leading technique for the discovery and identification of molecules from biological and environmental samples. However, the annotation of MS/MS spectra with molecular structures remains a challenge. Recently, we have introduced DreaMS – a neural network that converts each MS/MS spectrum into a high-dimensional vector encoding its molecular structure. By applying DreaMS to hundreds of millions of MS/MS spectra from diverse samples, such as human skin or blood, plant extracts, marine environments, food, and many others, we constructed DreaMS Atlas – a vast network that links diverse scientific studies and enables the interpretation of mass spectra through network analysis. The goal of this project is to make DreaMS Atlas accessible to the global community by developing an interactive web interface that facilitates network exploration and offers user-friendly features, such as querying the atlas with specific mass spectra or filtering it by organisms of interest. The ESM Metagenomic Atlas can serve as inspiration for this project.
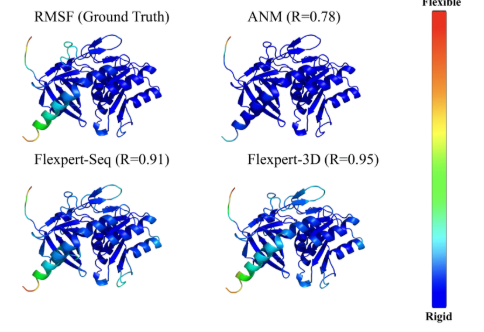
Generative models for design of dynamic proteins
Supervisors
Petr Kouba, Jiri Sedlar, Josef Sivic
Motivation
The domain of protein engineering and protein design is booming and is becoming a more and more prominent domain for applying machine learning. The scientific breakthroughs of machine learning achieved in protein design have been recognized by the 2024 Nobel Prize in chemistry, awarded to David Baker (U Washington), John Jumper, and Demis Hassabis (both DeepMind). The commercial potential of this interdisciplinary field is evidenced by many recently funded start-ups focusing on using ML-based protein design for the development of novel medicines, such as Xaira, Isomorphic Labs, or Prescient Design.

Generating „Realistic“ Camera Views of 3D Models
Supervisors
Torsten Sattler
Motivation
Modern deep-based computer vision approaches, e.g., local features, 3D reconstruction, etc., need large amounts of training data. Often, such methods need annotations that are expensive to obtain from real-world images, e.g., accurate scene geometry, pixel-level correspondences between images, depth maps, optical flow, etc. At the same time, large repositories of 3D models are becoming available, e.g., the 3D model sharing platform Sketchfab and 3D model datasets such as Objavers. This allows generating training images by rendering 3D models. This enables generating accurate annotations. This enables generating accurate annotations. However, this requires being able to identify a set of potentially interesting viewpoints per scene automatically in order to scale to many scenes.
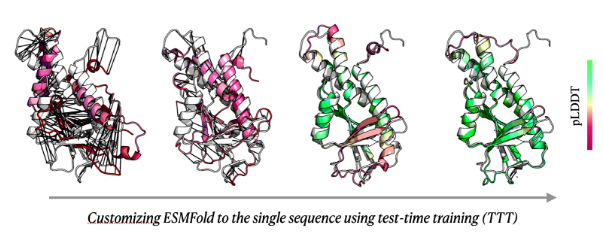
Customizing machine learning models for proteins
Supervisors
Anton Bushuiev, Roman Bushuiev, Jiri Sedlar, Josef Sivic
Motivation
Machine learning models often struggle with proteins and other biological data due to limited data and their high variability. Pretraining models on large datasets improves general performance but often sacrifices accuracy for individual proteins, even though biologists and other practitioners often focus on one specific protein they study at a time. Recently, we developed test-time training (TTT) for proteins, enabling the customization of machine learning models for one protein at a time and improving their accuracy in practical applications. While TTT provides a strong proof-of-concept for customizing machine learning models for proteins, several important directions remain underexplored. In this thesis, the student will explore more advanced customization techniques, leveraging methods previously used in computer vision and natural language processing, to develop a stronger approach with a significant impact on biological discoveries.
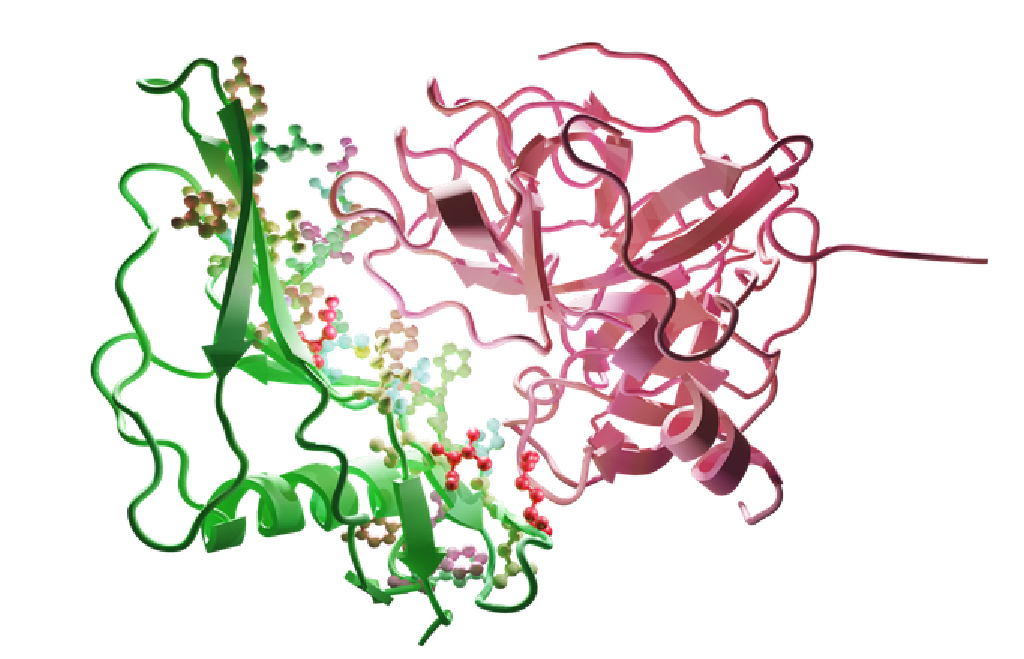
Machine learning for the design of protein-protein interactions
Supervisors
Anton Bushuiev, Roman Bushuiev, Petr Kouba, Jiri Sedlar, Jiri Damborsky, Stanislav Mazurenko, Josef Sivic
Motivation
Proteins are large molecules that drive nearly all processes in living cells. The analysis of protein-protein interactions (PPIs) and their design unlocks application areas of tremendous importance, most notably in healthcare and biotechnology2. Recently, we have developed PPIformer, a new self-supervised method to design protein-protein interactions by training from a large number of potentially all known PPI structures. PPIformer was shown to outperform existing methods and, therefore, opens up an exciting space for its further development and applications to practical case studies.